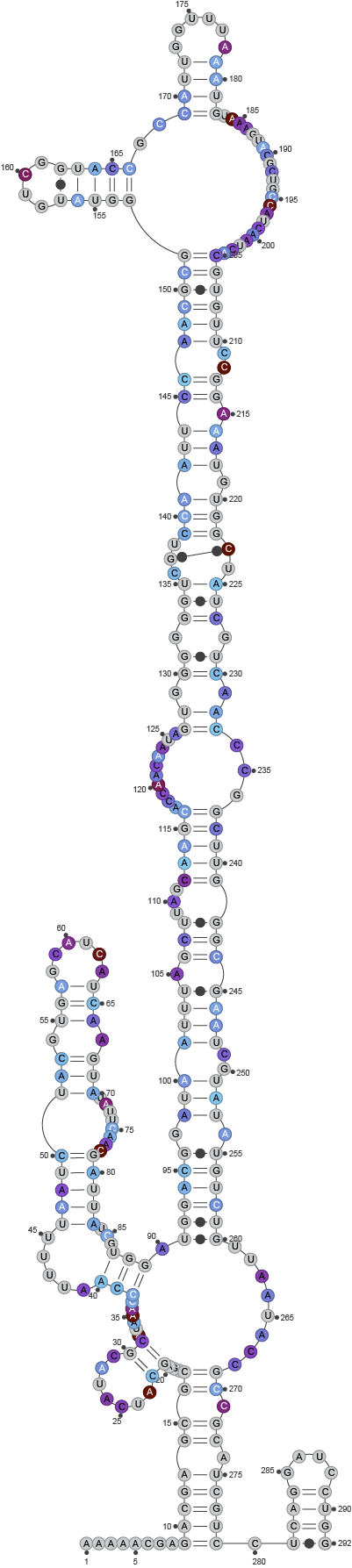
Example 4) Masking using a regions file
When working with a complex sequence, SEISMIC-RNA allows to select distinct parts of it for analyzing.
In the case that, for example, a pair of primers was used to create an amplicon, it is possible to mask the positions of the primers using a regions file (see Metadata for Regions). Conceptually, this prevents the lack of mutations in the primer sequence regions to be incorrectly considered low reactivity when folding.
Download example files
Paired-end FASTQ files were generated using seismic sim. The files, with
their corresponding reference fasta file and the regions file, can be
downloaded here:
https://raw.githubusercontent.com/rouskinlab/seismic-rna/main/src/userdocs/examples/MultiSample/fq/RegionsFile.zip
Run the SEISMIC-RNA workflow
The regions file can be included using --mask-regions-file. This will
mask the sequences determined by the file, and then use the rest for folding
(for which the flags --fold and --draw are added):
seismic wf fq/Regions_Ref.fa -x fq/ --mask-regions-file fq/regions_file.csv --fold --draw
Output
Given that the primer sequences were masked, the model shows no reactivity at the first nor last 20 bases: